Getting started with the API¶
Chemfp has an extensive API. This chapter gives examples of how to use the high-level commands for different types of similarity search, fingerprint generation, and MaxMin diversity picking.
Get ChEMBL 32 fingerprints in FPB format¶
Many of the examples in this section use the RDKit Morgan fingerprints for ChEMBL 32. These are available in FPS format from ChEMBL. I distributed a version in FPB format which includes a chemfp license key to unlock all chemfp functionality for that file.
You will need to download and uncompress that file. Here is one way:
% curl -O https://chemfp.com/datasets/chembl_32.fpb.gz
% gunzip chembl_32.fpb.gz
The FPB file includes the required ChEMBL notices. To see them use:
% chemfp fpb_text chembl_32.fpb
Similarity search of ChEMBL by id¶
In this section you’ll use the high-level subsearch command to find the closest neighbors to a named fingerprint record.
CHEMBL113 is the ChEMBL record for caffeine.
What are the 5 most similar fingerprints in ChEMBL 32?
Chemfp has several “high-level” functions which provide a lot of
functionality through a single function call. The
chemfp.simsearch() function, like the simsearch <simsearch>
command, implements similarity searches.
If given a query id and a target filename then it will open the target fingerprints, find the first fingerprint whose the id matches the query id, and use that fingerprint for a similarity search. When no search parameters are specified, it does a Tanimoto search for the 3 nearest neighbors, which in this case will include CHEMBL113 itself.
>>> import chemfp
>>> result = chemfp.simsearch(query_id="CHEMBL113", targets="chembl_32.fpb")
>>> result
SingleQuerySimsearch('3-nearest Tanimoto search. #queries=1, #targets=2327928
(search: 138.46 ms total: 177.29 ms)', result=SearchResult(#hits=3))
The SingleQuerySimsearch summaries the query and the search
times. In this case it took about 0.14 seconds for the actual
fingerprint search, and 0.18 seconds total; the high-level
organization also has some overhead.
Note
Timings are highly variable. It took 180 ms because the content was not in disk cache. Re-running with a warm cache took only 66 ms total.
The simsearch result object depends on the type of search. In this case it was a single query vs. a set of targets. The two other supported cases 1) multiple queries against a set of targets, and 2) using the same fingerprints for both targets and queries (a “NxN” search).
The result is a wrapper (really, a proxy) for the underlying search object, which you can get through result.result:
>>> result.result
SearchResult(#hits=3)
You can access the underlying object directly, but I found myself often forgetting to do that, so the simsearch result objects also implement the underlying search result(s) API, by forwarding them to the actual result, which lets you do things like:
>>> result.get_ids_and_scores()
[('CHEMBL113', 1.0), ('CHEMBL4591369', 0.7333333333333333), ('CHEMBL1232048', 0.7096774193548387)]
As expected, CHEMBL113 is 100% identical to itself. The next closest records are CHEMBL4591369 and CHEMBL1232048.
If you have Pandas installed then use SearchResult.to_pandas()
to export the target id and score to a Pandas Dataframe:
>>> result.to_pandas()
target_id score
0 CHEMBL113 1.000000
1 CHEMBL4591369 0.733333
2 CHEMBL1232048 0.709677
The k parameter specifies the number of neighbors to select. The following finds the 10 nearest neighbors and exports the results to a Pandas table using the columns “id” and “Tanimoto”:
>>> chemfp.simsearch(query_id="CHEMBL113",
... targets="chembl_32.fpb", k=10).to_pandas(columns=["id", "Tanimoto"])
id Tanimoto
0 CHEMBL113 1.000000
1 CHEMBL4591369 0.733333
2 CHEMBL1232048 0.709677
3 CHEMBL446784 0.677419
4 CHEMBL1738791 0.666667
5 CHEMBL2058173 0.666667
6 CHEMBL286680 0.647059
7 CHEMBL417654 0.647059
8 CHEMBL2058174 0.647059
9 CHEMBL1200569 0.641026
Similarity search of ChEMBL using a SMILES¶
In this section you’ll use the high-level subsearch command to find target fingerprints which are at leats 0.75 similarity to a given SMILES string.
Wikipedia gives O=C(NCc1cc(OC)c(O)cc1)CCCC/C=C/C(C)C as a SMILES for capsaicin. What ChEMBL structures are at leat 0.75 similar to that SMILES?
The chemfp.simsearch() function supports many types of query
inputs. The previous section used used a query id to look up the
fingerprint in the targets. Alternatively, the query parameter takes
a string containing a structure record, by default in “smi”
format. (Use query_format to specify “sdf”, “molfile”, or other
supported format.):
>>> import chemfp
>>> result = chemfp.simsearch(query = "O=C(NCc1cc(OC)c(O)cc1)CCCC/C=C/C(C)C",
... targets="chembl_32.fpb", threshold=0.75)
>>> len(result)
23
>>> result.to_pandas()
target_id score
0 CHEMBL4227443 0.791667
1 CHEMBL87171 0.795918
2 CHEMBL76903 0.764706
3 CHEMBL80637 0.764706
4 CHEMBL86356 0.764706
5 CHEMBL88024 0.764706
6 CHEMBL87024 0.764706
7 CHEMBL89699 0.764706
8 CHEMBL88913 0.764706
9 CHEMBL89176 0.764706
10 CHEMBL89356 0.764706
11 CHEMBL89829 0.764706
12 CHEMBL121925 0.764706
13 CHEMBL294199 1.000000
14 CHEMBL313971 1.000000
15 CHEMBL314776 0.764706
16 CHEMBL330020 0.764706
17 CHEMBL424244 0.764706
18 CHEMBL1672687 0.764706
19 CHEMBL1975693 0.764706
20 CHEMBL3187928 1.000000
21 CHEMBL88076 0.750000
22 CHEMBL313474 0.750000
The next section will show how to sort the results by score.
Sorting the search results¶
The previous section showed how to find all ChEMBL fingerprints with 0.75 similarity to capsaicin. This section shows how to sort the scores in chemfp, which is faster than using Pandas to sort.
While chemfp’s k-nearest search orders the hits from highest to lowest, the threshold search order is arbitrary (or more precisely, based on implementation-specific factors). If you want sorted scores in the Pandas table you could do so using Pandas’ sort_values method, as in the following:
>>> chemfp.simsearch(query = "O=C(NCc1cc(OC)c(O)cc1)CCCC/C=C/C(C)C",
... targets="chembl_32.fpb", threshold=0.75).to_pandas().sort_values("score", ascending=False)
target_id score
20 CHEMBL3187928 1.000000
14 CHEMBL313971 1.000000
13 CHEMBL294199 1.000000
1 CHEMBL87171 0.795918
0 CHEMBL4227443 0.791667
19 CHEMBL1975693 0.764706
18 CHEMBL1672687 0.764706
17 CHEMBL424244 0.764706
16 CHEMBL330020 0.764706
15 CHEMBL314776 0.764706
12 CHEMBL121925 0.764706
11 CHEMBL89829 0.764706
10 CHEMBL89356 0.764706
9 CHEMBL89176 0.764706
8 CHEMBL88913 0.764706
7 CHEMBL89699 0.764706
6 CHEMBL87024 0.764706
5 CHEMBL88024 0.764706
4 CHEMBL86356 0.764706
3 CHEMBL80637 0.764706
2 CHEMBL76903 0.764706
21 CHEMBL88076 0.750000
22 CHEMBL313474 0.750000
However, it’s a bit faster to have chemfp sort the results before exporting to Pandas. (For a large data set, about 10x faster.)
The simsearch reordering parameter takes one of the following strings:
- “increasing-score” - sort by increasing score
- “decreasing-score” - sort by decreasing score
- “increasing-score-plus” - sort by increasing score, break ties by increasing index
- “decreasing-score-plus” - sort by decreasing score, break ties by increasing index
- “increasing-index” - sort by increasing target index
- “decreasing-index” - sort by decreasing target index
- “move-closest-first” - move the hit with the highest score to the first position
- “reverse” - reverse the current ordering
Here’s how order the simsearch results by “decreasing-score”:
>>> chemfp.simsearch(query = "O=C(NCc1cc(OC)c(O)cc1)CCCC/C=C/C(C)C",
... targets="chembl_32.fpb", threshold=0.75, ordering="decreasing-score").to_pandas()
target_id score
0 CHEMBL294199 1.000000
1 CHEMBL313971 1.000000
2 CHEMBL3187928 1.000000
3 CHEMBL87171 0.795918
4 CHEMBL4227443 0.791667
5 CHEMBL76903 0.764706
6 CHEMBL80637 0.764706
7 CHEMBL86356 0.764706
8 CHEMBL88024 0.764706
9 CHEMBL87024 0.764706
10 CHEMBL89699 0.764706
11 CHEMBL88913 0.764706
12 CHEMBL89176 0.764706
13 CHEMBL89356 0.764706
14 CHEMBL89829 0.764706
15 CHEMBL121925 0.764706
16 CHEMBL314776 0.764706
17 CHEMBL330020 0.764706
18 CHEMBL424244 0.764706
19 CHEMBL1672687 0.764706
20 CHEMBL1975693 0.764706
21 CHEMBL88076 0.750000
22 CHEMBL313474 0.750000
Fingerprints as byte strings¶
In this section you’ll learn how chemfp uses byte strings to represent a fingerprint.
Chemfp works with binary fingerprints, typically in the range of a few hundred to a few thousand bits, and with a bit density high enough that sparse representations aren’t useful.
There are several common ways to represent these fingerprints. Chemfp uses a byte string, including at the Python level. (Two common alternatives are to have detected “fingerprint object”, or to re-use something like Python’s arbitrary-length integers.)
Here, for example, is a 24-bit fingerprint in Python:
>>> fp = b"XYZ"
The leading ‘b’ indicates the string is a byte string.
Remember, normal Python strings (also called Unicode strings) are different from byte strings. Byte strings are a sequence of 8-bits values, while normal strings use a more complicated representation that can handle the diversity of human language.
Chemfp includes functions in the bitops module to work with byte-string fingerprints. For example, here are the positions of the on-bits in the fingerprint:
>>> from chemfp import bitops
>>> bitops.byte_to_bitlist(b"XYZ")
[3, 4, 6, 8, 11, 12, 14, 17, 19, 20, 22]
>>> bitops.byte_to_bitlist(b"ABC")
[0, 6, 9, 14, 16, 17, 22]
and here is the Tanimoto between those two byte strings:
>>> bitops.byte_tanimoto(b"ABC", b"XYZ")
0.2857142857142857
You can verify they share bits 6, 14, 17, and 22, while the other 10 bits are distinct, giving a Tanimoto of 4/14 = 0.2857142857142857.
If you are working with hex-encoded fingerprints then you can use Python’s bytestring methods to convert to- and from- those values:
>>> b"ABC".hex()
'414243'
>>> bytes.fromhex("4101ff")
b'A\x01\xff'
Python’s fromhex() does not accept a byte string:
>>> bytes.fromhex(b"4101ff")
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
TypeError: fromhex() argument must be str, not bytes
If you have a byte string you might consider using chemfp’s
hex_decode() function, which supports both types of strings:
>>> bitops.hex_decode("4101ff")
b'A\x01\xff'
>>> bitops.hex_decode(b"4101ff")
b'A\x01\xff'
or use one of the “hex” functions in the bitops module, which work on hex-encoded fingerprints:
>>> bitops.hex_to_bitlist("4101ff")
[0, 6, 8, 16, 17, 18, 19, 20, 21, 22, 23]
>>> bitops.byte_to_bitlist(b"A\x01\xff")
[0, 6, 8, 16, 17, 18, 19, 20, 21, 22, 23]
Generating Fingerprints¶
In this section you’ll learn how to generate a fingerprint using chemfp’s interface to several supported cheminformatics toolkits.
Chemfp is a package for working with cheminformatics fingerprints. It is not a cheminformatics toolkit. It instead knows how to use OpenEye’s OEChem and OEGraphSim, the RDKit, Open Babel, and CDK to read and write molecules, and to generate fingerprints.
I’ll use the MACCS fingerprints in this section because they are a well-known fingerprint with a long history in this field, they implemented in all of the supported toolkits, and they are short enough to include in the documentation. However:
Important
Do not use the MACCS fingerprints in modern research. They are only of historical interest.
In chemfp, a FingerprintType describes an object which can
generate fingerprints, typically from a structure record. These know
what kind of fingerprints to generate (eg, “MACCS” or “circular”;
chemfp calls this a “fingerprint family”), as well as any
configuration parameters (eg, fingerprint size, radius, and atom type
flags).
Each of chemfp’s toolkit interfaces includes several FingerprintTypes, one for the toolkit’s fingerprint types, using the default parameters, if any. Here are the ones for the MACCS keys:
>>> import chemfp
>>> chemfp.rdkit.maccs166
RDKitMACCSFingerprintType_v2(<RDKit-MACCS166/2>)
>>> chemfp.openbabel.maccs
OpenBabelMACCSFingerprintType_v2(<OpenBabel-MACCS/2>)
>>> chemfp.cdk.maccs
CDKMACCSFingerprintType_v20(<CDK-MACCS/2.0>)
>>> chemfp.openeye.maccs166
OpenEyeMACCSFingerprintType_v3(<OpenEye-MACCS166/3>)
Chemfp’s choice of “maccs” vs “maccs166” follows the naming convention of the underlying toolkit.
The fingerprint types subclasses implement from_smiles which takes
a SMILES string, uses the underlying toolkit to parse the SMILES into
a toolkit molecule and generate a fingerprint, and returns the
fingerprint as a byte string.:
>>> import chemfp
>>> chemfp.rdkit.maccs166.from_smiles("Cn1cnc2c1c(=O)[nH]c(=O)n2C")
b'\x00\x00\x00\x000\x00\x00\x00\x01\xd4\x14\xd9\x13#\x91S\x80\xe1x\xea\x1f'
>>> chemfp.openbabel.maccs.from_smiles("Cn1cnc2c1c(=O)[nH]c(=O)n2C")
b'\x00\x00\x00\x000\x00\x00\x00\x01\xd4\x14\xd9\x13#\x91C\x80\xe1x\xea\x1f'
>>> chemfp.cdk.maccs.from_smiles("Cn1cnc2c1c(=O)[nH]c(=O)n2C")
b'\x00\x00\x00\x000\x00\x00\x00\x01\xd4\x14\xd9\x03#\x91C\x80\xe1x\xea\x1f'
>>> chemfp.openeye.maccs166.from_smiles("Cn1cnc2c1c(=O)[nH]c(=O)n2C")
b'\x00\x00\x00\x000\x00\x00\x00\x01\xd4\x14\xd9\x13\xa3\x91C\x80\xa0zj\x1b'
What’s the difference between the RDKit and Open Babel or CDK
fingerprints? I’ll use byte_to_bitlist() to get the bits in each
one, then use Python’s set algebra to see which bits are set in one but no the other:
>>> from chemfp import bitops
>>> set(bitops.byte_to_bitlist(rd_maccs)) - set(bitops.byte_to_bitlist(ob_maccs))
{124}
>>> set(bitops.byte_to_bitlist(ob_maccs)) - set(bitops.byte_to_bitlist(rd_maccs))
set()
>>> set(bitops.byte_to_bitlist(rd_maccs)) - set(bitops.byte_to_bitlist(cdk_maccs))
{124, 100}
>>> set(bitops.byte_to_bitlist(cdk_maccs)) - set(bitops.byte_to_bitlist(rd_maccs))
set()
Some toolkits store MACCS key 1 in bit 0 (the first bit) and others store key 1 in bit 1, leaving bit 0 empty. Chemfp’s standardizes all MACCS fingerprints to start at bit 0.
Similarity Search of ChEMBL by fingerprint¶
In this section you’ll learn how to search ChEMBL using a fingerprint byte string as the query.
ChEMBL generated the ChEBML fingerprints using RDKit’s Morgan fingerprints with the default settings. The corresponding chemfp fingerprint type string is:
RDKit-Morgan/1 radius=2 fpSize=2048 useFeatures=0 useChirality=0 useBondTypes=1
This is version 1 of the Morgan fingerprints (the version is because fingerprint generation algorithms may change), with a radius of 2. It generates 2048-bit fingerprints. The atom invariants were used, rather than the feature invariants, chirality was not included, and bond type were included.
If you know this type string, you can use chemfp’s
chemfp.get_fingerprint_type() function to parse the string and return
the appropriate FingerprintType object, then parse a SMILES and get
its fingerprint:
>>> import chemfp
>>> fptype = chemfp.get_fingerprint_type(
... "RDKit-Morgan/1 radius=2 fpSize=2048 useFeatures=0 useChirality=0 useBondTypes=1")
>>> fptype
RDKitMorganFingerprintType_v1(<RDKit-Morgan/1 radius=2 fpSize=2048 useFeatures=0 useChirality=0 useBondTypes=1>)
>>> fp = fptype.from_smiles("Cn1cnc2c1c(=O)[nH]c(=O)n2C")
>>> fp[:10] + b" ... " + fp[-10:]
b'\x00\x00\x00\x00\x02\x00\x00\x02\x00\x00 ... \x00\x00\x00\x00\x00\x00\x00\x00\x00\x00'
Alternatively you can use the default Morgan2 fingerprint type from
the rdkit_toolkit module, available through the
chemfp.rdkit helper:
>>> fp = chemfp.rdkit.morgan2.from_smiles("Cn1cnc2c1c(=O)[nH]c(=O)n2C")
Use query_fp to have chemfp.simsearch() search using the given
fingerprint as a query:
>>> chemfp.simsearch(query_fp=fp, targets="chembl_32.fpb", k=4).to_pandas()
target_id score
0 CHEMBL1114 1.000000
1 CHEMBL2106653 0.794118
2 CHEMBL131371 0.638889
3 CHEMBL2107523 0.613636
Loading fingerprints into an arena¶
In this section you’ll learn about chemfp’s fingerprint “arena”, and how to load one from a fingerprint file.
The fastest way for chemfp to do a similarity search is to load all of
the fingerprints in memory in a data struture called an “arena”. Use
the chemfp.load_fingerprints() command to read an FPS or FPB file into
an arena, like this:
>>> import chemfp
>>> arena1 = chemfp.load_fingerprints("chembl_32.fps.gz")
>>> arena2 = chemfp.load_fingerprints("chembl_32.fpb")
If you do those steps you’ll see arena1 takes about 10 seconds to load, while arena2 takes well less than a second.
There’s a very straight-forward explanation. Chemfp must decompress and parse the fps.gz file and arrange the content into an arena, while the fpb file is internally already structured as an arena, so requires little additional processing.
What’s in an arena? Here’s the Python representation of both arenas, and a few operations on arena2:
>>> arena1
FingerprintArena(#fingerprints=2327928)
>>> arena2
FPBFingerprintArena(#fingerprints=2327928)
>>> len(arena2)
2327928
>>> arena2.get_index_by_id("CHEMBL113")
40336
>>> arena2.ids[40336]
'CHEMBL113'
>>> arena2.fingerprints[40336][:16]
b'\x00\x00\x00\x00\x02\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00'
>>> arena2[40336]
('CHEMBL113', b'\x00\x00\x00\x00\x02\x00\x00\x00\x00\x00
\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00
\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00
... many lines removed ...
>>> arena2.fingerprints.random_choice()[:8]
b'\x02\x00\x00\x00\x00@\x00\x01'
The main difference between a FingerprintArena and an
FPBFingerprintArena is the FPBFingerprintArena uses a
memory-mapped to access the file contents, which means it as an open
file handle. Use close method or a context manager if you don’t want
to depend on Python’s garbage collector to close the file. (The
FingerprintArena also implements close, which does nothing.)
The FPS and FPB files may store the fingerprint type string as metadata, which is available from the arena:
>>> import chemfp
>>> arena = chemfp.load_fingerprints("chembl_32.fpb")
>>> arena.metadata.type
'RDKit-Morgan/1 radius=2 fpSize=2048 useFeatures=0 useChirality=0 useBondTypes=1'
Use the arena’s FingerprintReader.get_fingerprint_type() method
to get corresponding FingerprintType object:
>>> arena.get_fingerprint_type()
RDKitMorganFingerprintType_v1(<RDKit-Morgan/1 radius=2 fpSize=2048 useFeatures=0 useChirality=0 useBondTypes=1>)
Generate an NxN similarity matrix¶
In this section you’ll learn how to do an NxN search with simsearch(), using an arena as both the queries and the targets.
The simsearch examples in the previous sections all took an FPB filename for the targets, which means the function had to open the file each time. You can also use an arena as the targets, which is useful if you will be doing many searches.:
>>> import chemfp
>>> arena = chemfp.load_fingerprints("chembl_32.fpb")
>>> chemfp.simsearch(query_id="CHEMBL1114", targets=arena, k=2).to_pandas()
target_id score
0 CHEMBL1114 1.000000
1 CHEMBL2106653 0.794118
>>> chemfp.simsearch(query_id="CHEMBL2106653", targets=arena, k=2).to_pandas()
target_id score
0 CHEMBL2106653 1.000000
1 CHEMBL1114 0.794118
(This means theobromine and theobromine sodium acetate are both each other’s nearest neighbors.)
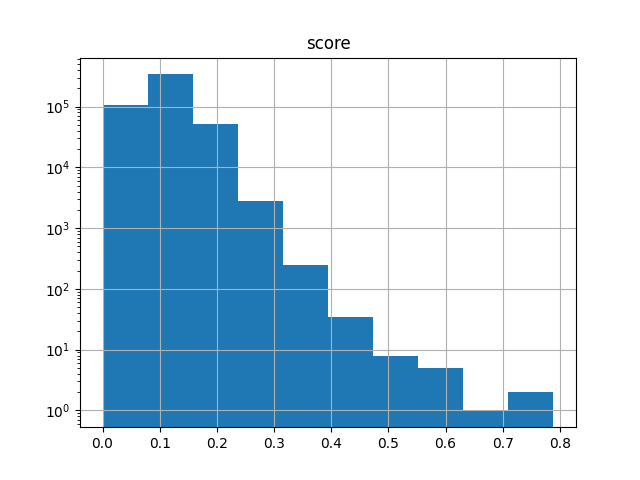
It’s also useful when working with subsets. The following selects 1000 fingerprints at random (using a specified seed for reproducibility), computes the N*(N-1)/2 upper-triangle similarity matrix of all pairwise comparisons, and generates a histogram of the scores:
>>> subset = arena.sample(1000, rng=87501)
>>> subset
FingerprintArena(#fingerprints=1000)
>>> ut = chemfp.simsearch(targets=subset, NxN=True,
... threshold=0.0, include_lower_triangle=False)
queries: 100%|█████████████████████| 1000/1000 [00:00<00:00, 57873.22 fps/s]
>>> from matplotlib import pyplot as plt
>>> ut.to_pandas().hist("score", log=True)
If you are in the Jupyter notebook then you should see the following image:
If you are in the traditional Python shell then to see the image you’ll also need to do:
>>> from matplotlib import pyplot as plt
>>> plt.show()
Note
chemfp’s NxN search is designed for a sparse result matrix. While it can certainly be used to compute all pairwise comparisons, it will not be as fast as an algorithm designed for that purpose.
The NxN search displays a progress bar by default. If you do not want a progress bar, either pass progress = False to simsearch, or globally disable the use of the default progress bar with:
>>> chemfp.set_default_progressbar(False)
To re-enable it, use:
>>> chemfp.set_default_progressbar(True)
Generate an NxM similarity matrix¶
In this section you’ll learn how to use multiple queries to search a
set of target fingerprints using chemfp.simsearch().
Earlier you learned several ways to search the ChEMBL fingerprints using a single query. If you have multiple queries then use the queries parameter to pass a query arena to simsearch.
For example, given two fingerprint sets A and B, which fingerprint in A is closest to a fingerprint in B?
To make it a bit more interesting, I’ll let A be the first 1,000 ChEMBL records and B be the last 1,000 ChEMBL records, sorted by id, on the assumption they are roughly time-ordered.
The identifiers are available from the id attribute, but these are either in popcount order (the default) or input order.
Tip
“Popcount”, which is short for “population count”, is the number of 1-bits in the fingerprint
Here are a few examples. They show that id is a list-like object, and that the fingerprints of the first and last identifiers have very different popcounts:
>>> import chemfp
>>> arena = chemfp.load_fingerprints("chembl_32.fpb")
>>> arena.ids
IdLookup(<672164601 bytes>, #4=2327928, #8=0, data_start=595958048, index_start=625581486)
>>> arena.ids[:2]
IdLookup(<672164601 bytes>, #4=2, #8=0, data_start=595958048, index_start=625581486)
>>> list(arena.ids[:2])
['CHEMBL17564', 'CHEMBL4300465']
>>> list(arena.ids[-2:])
['CHEMBL4764287', 'CHEMBL4795718']
>>>
>>> from chemfp import bitops
>>> bitops.byte_popcount(arena.get_fingerprint_by_id("CHEMBL17564"))
1
>>> bitops.byte_popcount(arena.get_fingerprint_by_id("CHEMBL4795718"))
238
We can’t simply sort by id because the default string sort places “CHEMBL10” before “CHEMBL2”. Instead, we need to remove the leading “CHEMBL”, convert the rest to an integer, and then do the sort.
I’ll start with a function to remove the first 6 characters and convert the rest to an integer:
def get_serial_number(s):
return int(s[6:])
I’ll use that to sort all of the identifiers by serial number:
>>> sorted_ids = sorted(arena.ids, key = get_serial_number)
>>> sorted_ids[:3]
['CHEMBL1', 'CHEMBL2', 'CHEMBL3']
>>> sorted_ids[-3:]
['CHEMBL5096101', 'CHEMBL5096102', 'CHEMBL5096103']
Seems to work!
Next, I’ll make subsets A and B using the
FingerprintArena.copy() method, which if passed a list of
indices will make a copy of only that subset:
>>> subset_A = arena.copy(
... indices=[arena.get_index_by_id(id) for id in sorted_ids[:1000]])
>>> subset_B = arena.copy(
... indices=[arena.get_index_by_id(id) for id in sorted_ids[-1000:]])
Now for the similarity search step. I’ll do a k=1 nearest-neighbor search for each fingerprint in A to find its closest match in B:
>>> result = chemfp.simsearch(queries=subset_A, targets=subset_B, k=1)
queries: 100%|████████████████████| 1000/1000 [00:00<00:00, 210261.88 fps/s]
>>> result
MultiQuerySimsearch('1-nearest Tanimoto search. #queries=1000, #targets=1000
(search: 25.41 ms total: 25.74 ms)', result=SearchResults(#queries=1000, #targets=1000))
There are a few ways to find the hit with the highest score. One uses standard Python, but I need to explain the details.
The MultiQuerySimsearch object, just like the underlying
SearchResults object, implements index lookup, so I’ll focus
on the first hit:
>>> first_hit = result[0]
>>> first_hit
SearchResult(#hits=1)
The SearchResult object knows the initial query id and has
ways to get the target indices, ids, and scores for the hits:
>>> first_hit.query_id
'CHEMBL1141'
>>> first_hit.get_ids_and_scores()
[('CHEMBL5095042', 0.02564102564102564)]
>>> first_hit.get_scores()
array('d', [0.02564102564102564])
With a slightly clumsy list comprehension, here’s the highest scoring pair:
>>> max((hits.get_scores()[0], hits.query_id, hits.get_ids()[0]) for hits in result)
(1.0, 'CHEMBL612', 'CHEMBL5095505')
Alternatively, use Pandas:
>>> result.to_pandas().nlargest(1, columns="score")
query_id target_id score
97 CHEMBL405 CHEMBL5095505 1.0
along with a quick check for ties:
>>> result.to_pandas().nlargest(5, columns="score")
query_id target_id score
97 CHEMBL405 CHEMBL5095505 1.000000
111 CHEMBL612 CHEMBL5095505 1.000000
387 CHEMBL651 CHEMBL5095396 0.969697
28 CHEMBL609 CHEMBL5095420 0.916667
770 CHEMBL549 CHEMBL5095175 0.777778
No wonder people enjoy using Pandas!
Before leaving this section, I’ll also show an advanced technique to get the fingerprint indices sorted by serial number, without doing the id-to-index lookup:
>>> serial_numbers = [get_serial_number(id) for id in arena.ids]
>>> sorted_indices = sorted(range(len(serial_numbers)),
... key = serial_numbers.__getitem__)
>>> arena.ids[sorted_indices[0]]
'CHEMBL1'
>>> arena.ids[sorted_indices[-1]]
'CHEMBL5096103'
>>> subset_A = arena.copy(indices=sorted_indices[:100])
>>> subset_B = arena.copy(indices=sorted_indices[-100:])
I’ll leave this one as a puzzler, which it was to me the first time I came across it.
Generating fingerprint files¶
In this section you’ll learn how to process a structure file to generate fingerprint files in FPS or FPB format.
Here’s a simple SMILES named “example.smi”:
c1ccccc1O phenol
CN1C=NC2=C1C(=O)N(C(=O)N2C)C caffeine
O water
I’ll use chemfp.rdk2fps() to process the file. By default it
generates Morgan fingerprints:
>>> import chemfp
>>> chemfp.rdkit2fps("example.smi", "example.fps")
example.smi: 100%|███████████| 63.0/63.0 [00:00<00:00, 4.19kbytes/s]
ConversionInfo("Converted structures from 'example.smi'. #input_records=3,
#output_records=3 (total: 15.66 ms)")
The function returns a ConversionInfo instance with details
about the number of records read and written, and the time needed.
Here’s what the resulting FPS file looks like:
% fold -w 70 example.fps
#FPS1
#num_bits=2048
#type=RDKit-Morgan/1 radius=2 fpSize=2048 useFeatures=0 useChirality=0 useBondTypes=1
#software=RDKit/2021.09.4 chemfp/4.1
#source=example.smi
#date=2023-05-16T18:11:52
0000000000000000020000000000000000000000000000000000000000000000000000
0000000000000000000000000020000000000000000000000000000000000000000000
0000000000000000000000000000000000000000000000020000000000008000000000
0000000000000000000000000000000000000000000000000000000000000001000000
0000000000000000008000000000000000000000000000000000000000000000100000
0000000000000000000000000000000000000000000000000004000000000000000000
0000000000000000400000000400000000000000000000000200000000000000000000
0000000000000000000000 phenol
0000000002000000000000000000000000000000000000000000000000000000000000
0000000004000000000000000400000100000000000080000000000001000000000000
1000000000000000000000040000000000000000000000000000080000000000000000
0000000000000000000000900000000000000000000000010000000200000000000000
0000000200000000000008000000000000040000000000080000000000040000100000
0002000000011000000000000000200000000000000000000000000000000000000000
0000010000000000000000000000000000000000000000000200000000000000000000
0000000000000000000000 caffeine
0000000000000000000000000000000000000000000000000000000000000000000000
0000000000000000000000000000000000000000000000000000000000000000000000
0000000000000000000000000000000000000000000000000000000040000000000000
0000000000000000000000000000000000000000000000000000000000000000000000
0000000000000000000000000000000000000000000000000000000000000000000000
0000000000000000000000000000000000000000000000000000000000000000000000
0000000000000000000000000000000000000000000000000000000000000000000000
0000000000000000000000 water
To generate an FPB file, change the final extension to “.fpb”:
>>> chemfp.rdkit2fps("example.smi", "example.fpb")
example.smi: 100%|███████████| 63.0/63.0 [00:00<00:00, 17.2kbytes/s]
ConversionInfo("Converted structures from 'example.smi'. #input_records=3, #output_records=3 (total: 6.98 ms)")
Generating fingerprints with an alternative type¶
In this section you’ll learn how to specify the fingerprint type to use when generating a fingerprint file.
The chemfp.rdkit2fps() function supports RDKit-specific
fingerprint parameters. In the following I’ll specify the output is
None, which tells chemfp to write to stdout, and have it generate
128-bit Morgan fingerprints with a radius of 3:
>>> info = chemfp.rdkit2fps("example.smi", None, progress=False, radius=3, fpSize=128)
#FPS1
#num_bits=128
#type=RDKit-Morgan/1 radius=3 fpSize=128 useFeatures=0 useChirality=0 useBondTypes=1
#software=RDKit/2021.09.4 chemfp/4.1
#source=example.smi
#date=2023-05-16T18:14:14
20800008808000000700420010020400 phenol
0b0601089310190400840a4010270807 caffeine
00004000000000000000000000000000 water
and then 128-bit torsion fingerprints:
>>> info = chemfp.rdkit2fps("example.smi", None, progress=False, type="Torsion", fpSize=128)
#FPS1
#num_bits=128
#type=RDKit-Torsion/2 fpSize=128 targetSize=4
#software=RDKit/2021.09.4 chemfp/4.1
#source=example.smi
#date=2023-05-16T18:14:38
03300000000003000000000000000030 phenol
03130000001003001317000003300333 caffeine
00000000000000000000000000000000 water
The chemfp.ob2fps(), chemfp.oe2fps(), and
chemfp.cdk2fps() functions are the equivalents for Open Babel,
OpenEye, and CDK, respectively, like this example with Open Babel:
>>> info = chemfp.ob2fps("example.smi", None, progress=False)
#FPS1
#num_bits=1021
#type=OpenBabel-FP2/1
#software=OpenBabel/3.1.0 chemfp/4.1
#source=example.smi
#date=2023-05-16T18:15:01
0000000000000000000002000000000000000000000000000000000000
0000000000000000000000000000080000000000000200000000000000
0000000000000800000000000000000000000200000000800000000000
0040080000000000000000000000000002000000000000000000020000
000000200800000000000000 phenol
80020000000000c000004600004000000000040000000060000020c000
0004000400300041008021400000100001008000000000000040000080
0800000080000c000c00800028000000400000900000000000000c8000
0ca000008000014006000000000080000000c000004000080000070102
00000800001880002a000000 caffeine
0000000000000000000000000000000000000000000000000000000000
0000000000000000000000000000000000000000000000000000000000
0000000000000000000000000000000000000000000000000000000000
0000000000000000000000000000000000000000000000000000000000
000000000000000000000000 water
>>> info = chemfp.ob2fps("example.smi", None, progress=False, type="ECFP4", nBits=128)
#FPS1
#num_bits=128
#type=OpenBabel-ECFP4/1 nBits=128
#software=OpenBabel/3.1.0 chemfp/4.1
#source=example.smi
#date=2023-05-16T18:16:06
00580020020a00000c40000000200000 phenol
020c464800040488160400822920b040 caffeine
40020000000000000000000000000080 water
Alternatively, if you have a FingerprintType object or
fingerprint type string then you can use chemfp.convert2fps(),
as with these examples using CDK to generate ECFP2 fingerprints:
>>> info = chemfp.convert2fps("example.smi", None, type=chemfp.cdk.ecfp2(size=128), progress=False)
#FPS1
#num_bits=128
#type=CDK-ECFP2/2.0 size=128 perceiveStereochemistry=0
#software=CDK/2.8 chemfp/4.1
#source=example.smi
#date=2023-05-16T18:18:35
20000000280000008000000040000280 phenol
200a1000202008c08401000400001008 caffeine
00000000800000000000000000000000 water
>>> info = chemfp.convert2fps ("example.smi", None, type="CDK-ECFP2/2.0 size=256", progress=False)
#FPS1
#num_bits=256
#type=CDK-ECFP2/2.0 size=256 perceiveStereochemistry=0
#software=CDK/2.8 chemfp/4.1
#source=example.smi
#date=2023-05-16T18:18:54
0000000020000000000000000000028020000000080000008000000040000000 phenol
000a0000000000c0040100040000000020001000202008008001000000001008 caffeine
0000000080000000000000000000000000000000000000000000000000000000 water
Extracting fingerprints from SDF tags¶
In this section you’ll learn about the chemfp.sdf2fps() function to
extract and decode fingerprints stored as tags in an SD file.
Sometimes fingerprints are encoded as tag data in an SD file.
For example, PubChem distributes the PubChem fingerprints in tag data which looks like the following:
> <PUBCHEM_CACTVS_SUBSKEYS>
AAADceB7sAAAAAAAAAAAAAAAAAAAAAAAAAA8YIAABYAAAACx9AAAHgAQAAAADCjBngQ8
wPLIEACoAzV3VACCgCA1AiAI2KG4ZNgIYPrA1fGUJYhglgDIyccci4COAAAAAAQCAAAA
AAAACAQAAAAAAAAAAA==
(I’ve folded the fingerprint from one line to three lines to fit in the documentation page.)
Use the sdf2fps function to extract these sorts of fingerprints and generate an FPS or FPB file.
The pubchem flags configures the function to extract the PubChem fingerprints, like this:
>>> chemfp.sdf2fps("Compound_099000001_099500000.sdf.gz",
... "Compound_099000001_099500000.fps", pubchem=True)
Compound_099000001_099500000.sdf.gz: 100%|█████████████████| 6.77M/6.77M [00:00<00:00, 33.4Mbytes/s]
ConversionInfo("Converted SDF records from 'Compound_099000001_099500000.sdf.gz'.
#input_records=10740, #output_records=10740 (total: 207.39 ms)")
Among other things, this sets the fp_tag to “PUBCHEM_CACTVS_SUBSKEYS”, which tells sdf2fps to look for the fingerprint under that tag, and it tries to parse that tag value using the “cactvs” decoder. These could be set directly, as with the following:
>>> chemfp.sdf2fps("Compound_099000001_099500000.sdf.gz",
... "Compound_099000001_099500000.fps",
... fp_tag="PUBCHEM_CACTVS_SUBSKEYS", decoder="cactvs")
Fingerprints may be encoded in many different ways, and chemfp
supports many of them through function in chemfp.encodings:
>>> from chemfp import encodings
>>> encodings.<tab>
encodings.binascii encodings.from_hex(
encodings.from_base64( encodings.from_hex_lsb(
encodings.from_binary_lsb( encodings.from_hex_msb(
encodings.from_binary_msb( encodings.from_on_bit_positions(
encodings.from_cactvs( encodings.import_decoder(
encodings.from_daylight(
The “lsb” and “msb” mean “least-significant bit” and “most-significant-bit” order, because sometimes bits are counted from the left, and sometimes from the right.
Select diverse fingerprints with MaxMin¶
In this section you’ll learn how to use the chemfp.maxmin()
function to select diverse fingerprints from the ChEMBL fingerprints.
Similarity search looks for nearest neighbors. A dissimilarity search looks for dissimilar fingerprints. In chemfp, the dissimilarity score is defined as 1 - Tanimoto score.
Dissimilarity search is often used to select diverse fingerprints, for example, to identify outliers in a fingerprint data set, to reduce clumpiness in random sampling, or to pick compounds which are more likely to improve coverage of chemistry space.
There are many methods for diversity selection. Perhaps the most common is MaxMin, which is a iterative hueristic (i.e., approximate method) to select fingerprints which are the most dissimilar from each other.
The basic algorithm is straight-forward:
- Start with one or more picks;
- Of the remaining candidates, find the fingerprint most dissimilar from the picks;
- Repeat until done.
This is an approximate method because there are likely many fingerprints with the same dissimilarity score, and there’s no way to identify which of those is the globally next most dissimilar.
On the other hand, the MaxMin algorithm is fast - it does not start by computing the full similarity matrix and and start returning picks right away, and these picks are often good enough.
Let’s give it a go and pick 100 diverse compounds from ChEMBL!:
>>> import chemfp
>>> picks = chemfp.maxmin("chembl_32.fpb", num_picks=100)
picks: 100%|█████████████████████████████| 100/100 [00:07<00:00, 13.95/s]
>>> picks
MaxMinScoreSearch('picked 100 fps. similarity <= 1.0,
#candidates=2327928, seed=1986293869 (pick: 7.17 s, total: 7.20 s)',
picker=MaxMinPicker(#candidates=2327828, #picks=100),
result=PicksAndScores(#picks=100))
There’s a lot in that message, but I think you can figure out most of
it. If you don’t pass in your own seed then it uses Python’s RNG to
get a seed, which is saved for the reproducibility’s sake. The picker
is the MaxMinPicker used, which is available for additional
picks. The results contain the picks and corresponding Tanimoto scores
(note: NOT dissimilarity score).
You can access and use the result object directly, but I found myself
often forgetting to do that, so the maxmin result objects -
MaxMinScoreSearch in this case - also implement the
underlying result API, by forwarding them to the actual result, which
lets you do things like:
>>> picks.get_scores()
[0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0,
0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0,
0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0,
0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0,
0.0, 0.0, 0.0, 0.0, 0.009259259259259259, 0.011111111111111112,
0.011111111111111112, 0.011235955056179775, 0.011235955056179775,
0.011235955056179775, 0.021739130434782608, 0.022222222222222223,
0.024390243902439025, 0.02857142857142857, 0.034482758620689655,
0.035398230088495575, 0.03571428571428571, 0.03636363636363636,
0.037037037037037035, 0.037037037037037035, 0.04225352112676056,
0.042735042735042736, 0.044444444444444446, 0.045454545454545456,
0.047619047619047616, 0.048, 0.04827586206896552,
0.049019607843137254, 0.04950495049504951, 0.05, 0.05, 0.05,
0.05454545454545454, 0.05555555555555555, 0.05555555555555555,
0.056338028169014086, 0.05660377358490566, 0.058823529411764705,
0.058823529411764705, 0.058823529411764705, 0.06097560975609756,
0.061224489795918366, 0.06153846153846154, 0.0625, 0.0625,
0.06349206349206349, 0.06521739130434782, 0.06521739130434782]
>>> sum(1 for score in picks.get_scores() if score != 0)
44
>>> df = picks.to_pandas()
>>> df.tail()
pick_id score
95 CHEMBL2096633 0.062500
96 CHEMBL9081 0.062500
97 CHEMBL3480491 0.063492
98 CHEMBL76815 0.065217
99 CHEMBL1688701 0.065217
>>>
>>> df.query("score > 0.0").head()
pick_id score
56 CHEMBL117334 0.009259
57 CHEMBL3186904 0.011111
58 CHEMBL25340 0.011111
59 CHEMBL1200706 0.011236
60 CHEMBL3188292 0.011236
I’ll ask the picker for the next 5 picks, first, without scores:
>>> picks.picker.pick_n(5).to_pandas()
pick_id
0 CHEMBL4644504
1 CHEMBL1985075
2 CHEMBL1765721
3 CHEMBL3985174
4 CHEMBL4979988
and then with scores:
>>> picks.picker.pick_n_with_scores(5).to_pandas()
pick_id score
0 CHEMBL1968682 0.068966
1 CHEMBL3484651 0.069231
2 CHEMBL4869925 0.069307
3 CHEMBL376289 0.069767
4 CHEMBL1869503 0.070423
If no initial fingerprint is specified then maxmin by default uses the heapsweep algorithm to find the globally most dissimilar fingerprint as the initial pick.
If you have a specific fingerprint in mind you can pass its index or id in as the initial seed:
>>> chemfp.maxmin("chembl_32.fpb", initial_pick="CHEMBL113", num_picks=3).to_pandas()
picks: 100%|█████████████████████████████████| 3/3 [00:00<00:00, 11.38/s]
pick_id score
0 CHEMBL113 0.0
1 CHEMBL4583200 0.0
2 CHEMBL1887585 0.0
>>>
>>> arena = chemfp.load_fingerprints("chembl_32.fpb")
>>> arena.get_index_by_id("CHEMBL113")
40336
>>> chemfp.maxmin(arena, initial_pick=40336, num_picks=3).to_pandas()
picks: 100%|█████████████████████████████████| 3/3 [00:00<00:00, 11.74/s]
pick_id score
0 CHEMBL113 0.0
1 CHEMBL1370850 0.0
2 CHEMBL1200391 0.0
Use MaxMin with references¶
In this section you’ll learn how to use maxmin to pick candidates which most increase the diversity given a set of references.
For example, a vendor might offer 30 million compounds (“the candidates”), and you want to pick the ones which improve the diversity coverage of your in-house compond collection of 5 million compounds (“the references”) by purchasing 1,000 of the compounds.
I don’t have those available for you to download and test, so I would work through that specific example.
Instead, I’ll see which 10 fingerprints in ChEMBL 32 (“the candidates”) added the most diversity to ChEMBL 31 (“the references”).
For this example you’ll need to download both ChEMBL 32 (from above) and ChEMBL 31:
% curl -O https://chemfp.com/datasets/chembl_32.fpb.gz
% gunzip chembl_32.fpb.gz
% curl -O https://chemfp.com/datasets/chembl_31.fpb.gz
% gunzip chembl_31.fpb.gz
then use MaxMin with both candidates and references:
>>> import chemfp
>>> picks = chemfp.maxmin(candidates="chembl_32.fpb",
references="chembl_31.fpb", num_picks=10)
picks: 100%|█████████████████████████████████| 10/10 [02:14<00:00, 13.50s/]
that took 135 seconds to show the most diverse compounds are:
pick_id score
0 CHEMBL2111187 0.166667
1 CHEMBL2107816 0.181818
2 CHEMBL2052012 0.232877
3 CHEMBL2105800 0.250000
4 CHEMBL1232306 0.250000
5 CHEMBL5070220 0.260870
6 CHEMBL5076690 0.261538
7 CHEMBL5087625 0.264368
8 CHEMBL5077931 0.266055
9 CHEMBL5082060 0.280488
CHEMBL2111187 is “RADIUM DICHLORIDE”. CHEMBL2107816 is “RADIUM RA 223 DICHLORIDE”, and CHEMBL2052012 is “[3H]-CHLOROQUINE”.
Select diverse fingerprints with Heapsweep¶
At some point I’ll an an example using chemfp.heapsweep().
Until then, see the description in What’s new in chemfp 4.0.
Sphere Exclusion¶
At some point I’ll an an example using chemfp.spherex(). Until
then, see the description in What’s new in chemfp 4.0.
Directed Sphere Exclusion¶
At some point I’ll an an example using chemfp.spherex() with
ranks. Until then, see the description in What’s new in chemfp 4.0.
Butina clustering¶
At some point I’ll an an example using chemfp.butina(). Until
then, see the description in What’s new in chemfp 4.1.